Reference



OpenAI's Answer
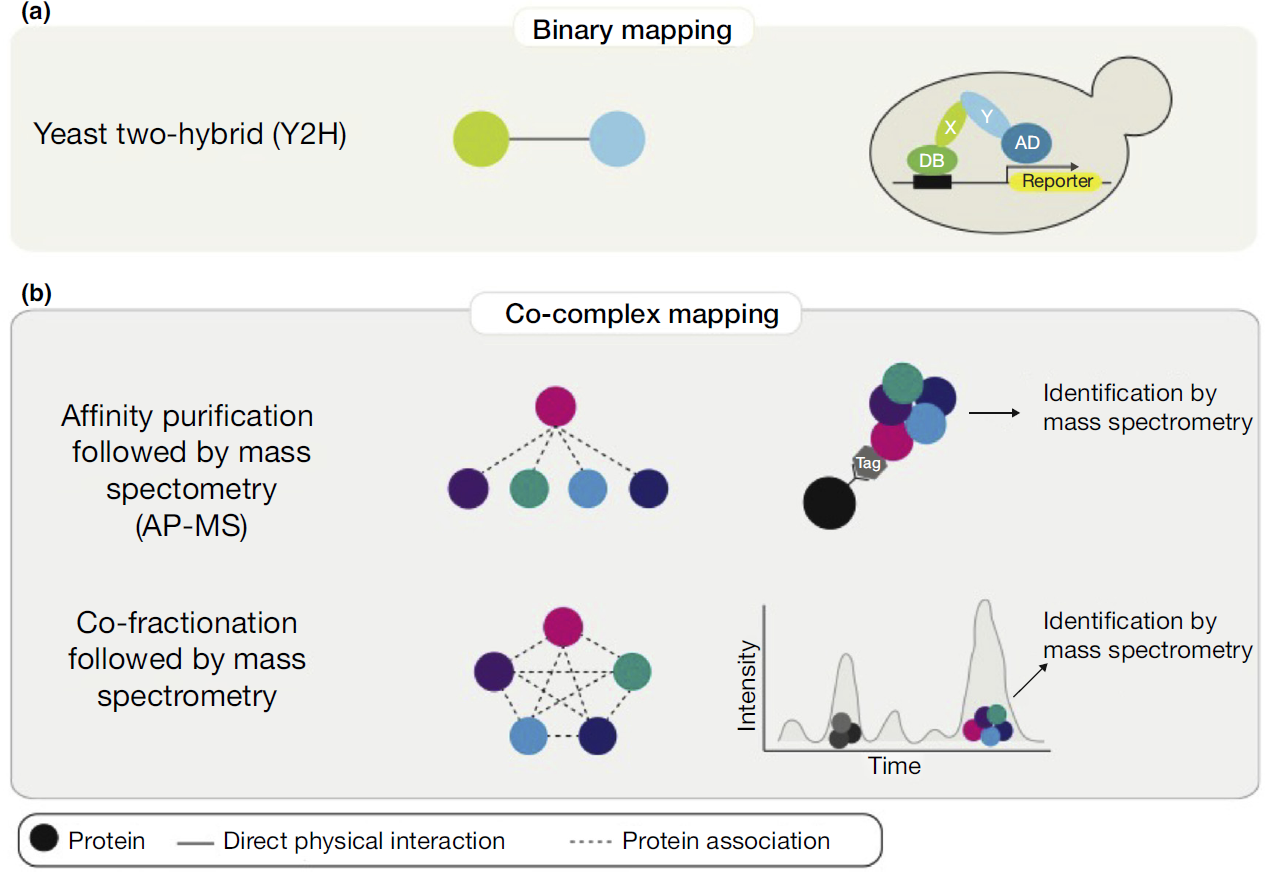
Large-scale mapping of human protein-protein interactions (PPIs) using mass spectrometry involves several advanced techniques. Here are some key methodologies used for this purpose:
-
Affinity Purification Mass Spectrometry (AP-MS):
- This technique involves the purification of protein complexes using specific antibodies followed by mass spectrometry to identify the components. It is widely used due to its ability to maintain the physiological context of PPIs.
- More about AP-MS can be found here.
-
Cross-linking Mass Spectrometry (XL-MS):
- XL-MS involves chemically cross-linking interacting proteins or protein regions in vivo, which can then be identified and analyzed using mass spectrometry. It is particularly useful for capturing transient or weak interactions.
- Learn more about XL-MS techniques.
-
Proximity-labeling Techniques:
- Methods like BioID and APEX use enzymes that label neighboring proteins within a certain radius, which can then be identified by mass spectrometry. These methods help in studying proteins that are in close proximity, possibly interacting within the cellular space.
-
Tandem Mass Tag (TMT) Labeling:
- TMT-based quantification allows multiplexed quantitative mass spectrometry, providing high throughput capability, essential for large-scale interactome studies.
-
Co-fractionation Mass Spectrometry:
- This involves fractionating cell lysates and analyzing the fractions with mass spectrometry. The co-migration of proteins across fractions can indicate protein interactions.

Each method provides unique insights and, when combined, delivers a comprehensive understanding of protein interaction networks and functionalities.
Follow Up
Related
What are the Catholic charities affiliated with the Archdiocese of New York?
How does Emma plan to use Mr. Gold to pull Excalibur from the stone?
How did Samuel Davis encourage the Daniell artists to explore the Himalayas?
What influence does Frank O'Hara's poetics have on Dmitry Kuzmin's poetry?
What role did Nicolas Tindal play in translating Paul de Rapin's History of England?
How many international caps did Patsy Gallacher earn for Ireland?